Curve Fitting: Linearization of Nonlinear Relationships
Linearization of Nonlinear Relationships
In the previous two sections, the model function ![]() was formed as a linear combination of functions
was formed as a linear combination of functions ![]() and the minimization of the sum of the squares of the differences between the model prediction and the data produced a linear system of equations to solve for the coefficients in the model. In that case
and the minimization of the sum of the squares of the differences between the model prediction and the data produced a linear system of equations to solve for the coefficients in the model. In that case ![]() was linear in the coefficients. In certain situations, it is possible to convert nonlinear relationships to a linear form similar to the previous methods. For example, consider the following models
was linear in the coefficients. In certain situations, it is possible to convert nonlinear relationships to a linear form similar to the previous methods. For example, consider the following models ![]() ,
, ![]() , and
, and ![]() :
:
![]()
![]() is an exponential model,
is an exponential model, ![]() is a power model, while
is a power model, while ![]() is a logarithmic model. These models are nonlinear in
is a logarithmic model. These models are nonlinear in ![]() and the unknown coefficients. However, by taking the natural logarithm of the first two, they can easily be transformed into linear models as follows:
and the unknown coefficients. However, by taking the natural logarithm of the first two, they can easily be transformed into linear models as follows:
![]()
In the first model, the data can be converted to ![]() and linear regression can be used to find the coefficients
and linear regression can be used to find the coefficients ![]() and
and ![]() . For the second model, the data can be converted to
. For the second model, the data can be converted to ![]() and linear regression can be used to find the coefficients
and linear regression can be used to find the coefficients ![]() , and
, and ![]() . The third model can be considered linear after converting the data into the form
. The third model can be considered linear after converting the data into the form ![]() .
.
Coefficient of Determination for Nonlinear Relationships
For nonlinear relationships, the coefficient of determination is not a very good measure for how well the data fit the model. See for example this article on the subject. In fact, different software will give different values for ![]() . We will use the coefficient of determination for nonlinear relationships defined as:
. We will use the coefficient of determination for nonlinear relationships defined as:
![]()
which is equal to 1 minus the ratio between the model sum of squares and the total sum of squares of the data. This is consistent with the definition of ![]() used in Mathematica for nonlinear models.
used in Mathematica for nonlinear models.
Example 1
Fit an exponential model to the data: (1,1.93),(1.1,1.61),(1.2,2.27),(1.3,3.19),(1.4,3.19),(1.5,3.71),(1.6,4.29),(1.7,4.95),(1.8,6.07),(1.9,7.48),(2,8.72),(2.1,9.34),(2.2,11.62).
Solution
The exponential model has the form:
![]()
This form can be linearized as follows:
![]()
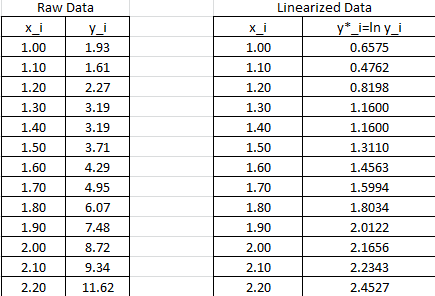
The data needs to be converted to ![]() .
. ![]() will be used to designate
will be used to designate ![]() . The following Microsoft Excel table shows the raw data, and after conversion to
. The following Microsoft Excel table shows the raw data, and after conversion to ![]() .
.
The linear regression described above will be used to find the best fit for the model:
![]()
with
![Rendered by QuickLaTeX.com \[\begin{split}a^*&=\frac{n\sum_{i=1}^nx_iy^*_i-\sum_{i=1}^nx_i\sum_{i=1}^ny^*_i}{n\sum_{i=1}^nx_i^2-\left(\sum_{i=1}^nx_i\right)^2}\\b^*&=\frac{\sum_{i=1}^ny^*_i-a^*\sum_{i=1}^nx_i}{n}\end{split}\]](https://engcourses-uofa.ca/wp-content/ql-cache/quicklatex.com-4454844df4d26460ac458e3771463de2_l3.png)
The following Microsoft Excel table is used to calculate the various entries in the above equation:
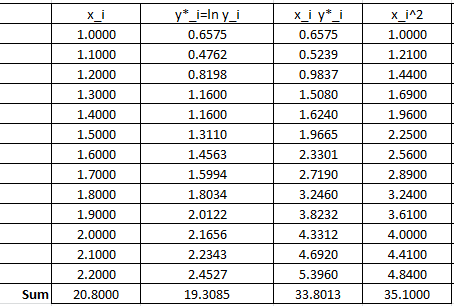
Therefore:
![Rendered by QuickLaTeX.com \[\begin{split}a^*&=\frac{13\times 33.8013-20.8\times 19.3085}{13\times 35.10-\left(20.8\right)^2}=1.5976\\b^*&=\frac{19.3085-1.5976\times 20.8}{13}=-1.0709\end{split}\]](https://engcourses-uofa.ca/wp-content/ql-cache/quicklatex.com-8becaa122aea6f02438419d4931f700c_l3.png)
These can be used to calculate the coefficients in the original model:
![]()
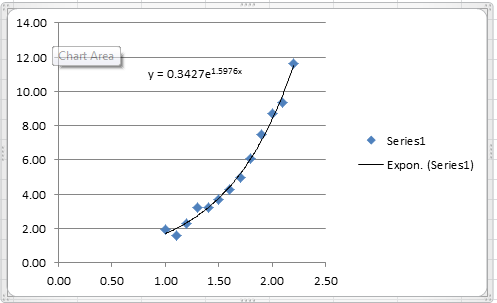
Therefore, the best exponential model based on the least squares of the linearized version has the form:
![]()
The following Microsoft Excel chart shows the calculated trendline in Excel with the same coefficients:
It is possible to calculate the coefficient of determination for the linearized version of this model, however, it would only describe how good the linearized model is. For the nonlinear model, we will use the coefficient of determination as described above which requires the following Microsoft Excel table: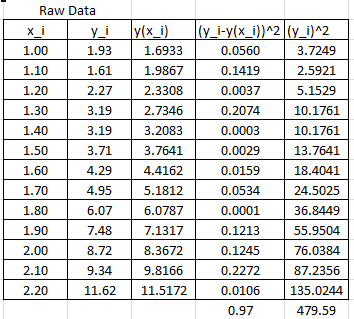
In this case, the coefficient of determination can be calculated as:
![]()
The NonlinearModelFit built-in function in Mathematica can be used to generate the model and calculate its ![]() as shown in the code below.
as shown in the code below.
Data = {{1, 1.93}, {1.1, 1.61}, {1.2, 2.27}, {1.3, 3.19}, {1.4, 3.19}, {1.5, 3.71}, {1.6, 4.29}, {1.7, 4.95}, {1.8, 6.07}, {1.9, 7.48}, {2, 8.72}, {2.1, 9.34}, {2.2, 11.62}};
model = NonlinearModelFit[Data, b1*E^(a1*x), {a1, b1}, x]
y = Normal[model]
R2 = model["RSquared"]
Plot[y, {x, 1, 2.2}, Epilog -> {PointSize[Large], Point[Data]}, PlotLegends -> {"Model"}, AxesLabel -> {"x", "y"}, AxesOrigin -> {0, 0} ]
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
Data = [[1, 1.93], [1.1, 1.61], [1.2, 2.27], [1.3, 3.19], [1.4, 3.19], [1.5, 3.71], [1.6, 4.29], [1.7, 4.95], [1.8, 6.07], [1.9, 7.48], [2, 8.72], [2.1, 9.34], [2.2, 11.62]]
def f(x, a, b): return a*np.exp(b*x)
coeff, covariance = curve_fit(f, [point[0] for point in Data],
[point[1] for point in Data])
print("coeff: ",coeff)
x_val = np.arange(1,2.2,0.01)
plt.title('%.5fe**(%.5fx)' % tuple(coeff))
plt.plot(x_val, f(x_val, coeff[0], coeff[1]))
plt.scatter([point[0] for point in Data], [point[1] for point in Data], c='k')
plt.xlabel("x"); plt.ylabel("y")
plt.grid(); plt.show()
# R squared
x = np.array([point[0] for point in Data])
y = np.array([point[1] for point in Data])
y_fit = f(x, coeff[0], coeff[1])
ss_res = np.sum((y - y_fit)**2)
ss_tot = np.sum((y - np.mean(y))**2)
r2 = 1 - (ss_res / ss_tot)
print("R Squared: ",r2)
The following link provides the MATLAB codes for implementing the Linearization of nonlinear exponential model.
Example 2
Fit a power model to the data: (1,1.93),(1.1,1.61),(1.2,2.27),(1.3,3.19),(1.4,3.19),(1.5,3.71),(1.6,4.29),(1.7,4.95),(1.8,6.07),(1.9,7.48),(2,8.72),(2.1,9.34),(2.2,11.62).
Solution
The power model has the form:
![]()
This form can be linearized as follows:
![]()
The data needs to be converted to ![]() .
. ![]() and
and ![]() will be used to designate
will be used to designate ![]() and
and ![]() respectively. The following Microsoft Excel table shows the raw data, and after conversion to
respectively. The following Microsoft Excel table shows the raw data, and after conversion to ![]() .
.
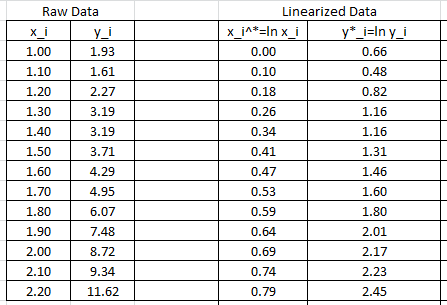
The linear regression described above will be used to find the best fit for the model:
![]()
with
![Rendered by QuickLaTeX.com \[\begin{split}a^*&=\frac{n\sum_{i=1}^nx_i^*y^*_i-\sum_{i=1}^nx_i^*\sum_{i=1}^ny^*_i}{n\sum_{i=1}^n(x_i^*)^2-\left(\sum_{i=1}^nx_i^*\right)^2}\\b^*&=\frac{\sum_{i=1}^ny^*_i-a^*\sum_{i=1}^nx_i^*}{n}\end{split}\]](https://engcourses-uofa.ca/wp-content/ql-cache/quicklatex.com-8d1ddaba26cba0cdea7f406f7c98a6ad_l3.png)
The following Microsoft Excel table is used to calculate the various entries in the above equation:
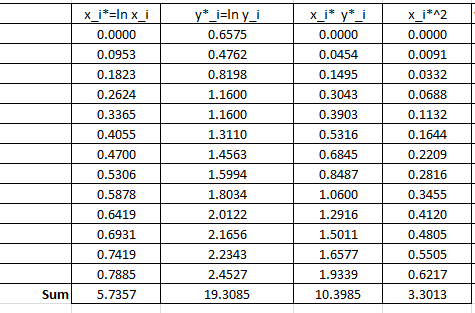
Therefore:
![Rendered by QuickLaTeX.com \[\begin{split}a^*&=\frac{13\times 10.3985-5.7357\times 19.3085}{13\times 3.3013-\left(5.7357\right)^2}=2.4387\\b^*&=\frac{19.3085-2.4387\times 5.7357}{13}=0.4093\end{split}\]](https://engcourses-uofa.ca/wp-content/ql-cache/quicklatex.com-3890aae29708ed93b655b772b816e4b3_l3.png)
These can be used to calculate the coefficients in the original model:
![]()
Therefore, the best power model based on the least squares of the linearized version has the form:
![]()
The following Microsoft Excel chart shows the calculated trendline in Excel with the same coefficients:
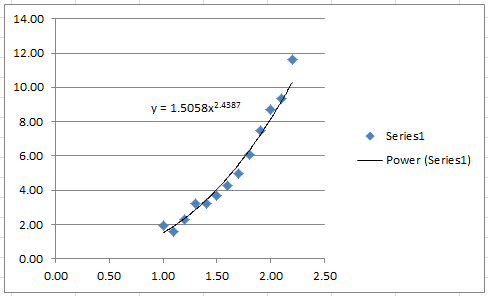
It is possible to calculate the coefficient of determination for the linearized version of this model, however, it would only describe how good the linearized model is. For the nonlinear model, we will use the coefficient of determination as described above which requires the following Microsoft Excel table: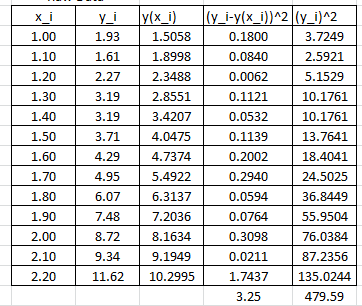
In this case, the coefficient of determination can be calculated as:
![]()
The NonlinearModelFit built-in function in Mathematica can be used to generate a slightly better model with a higher ![]() . The following is the corresponding Mathematica output.
. The following is the corresponding Mathematica output.
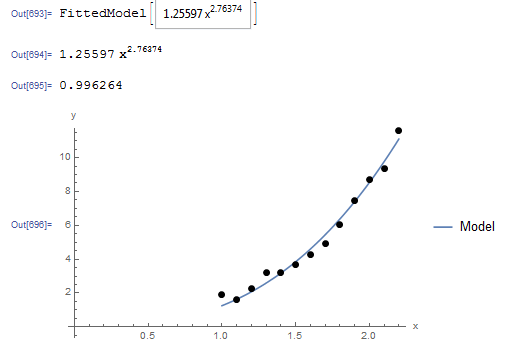
The Mathematica code is shown below.
View Mathematica CodeData = {{1, 1.93}, {1.1, 1.61}, {1.2, 2.27}, {1.3, 3.19}, {1.4, 3.19}, {1.5, 3.71}, {1.6, 4.29}, {1.7, 4.95}, {1.8, 6.07}, {1.9, 7.48}, {2, 8.72}, {2.1, 9.34}, {2.2, 11.62}};
model = NonlinearModelFit[Data, b1*x^(a1), {a1, b1}, x]
y = Normal[model]
R2 = model["RSquared"]
Plot[y, {x, 1, 2.2}, Epilog -> {PointSize[Large], Point[Data]}, PlotLegends -> {"Model"}, AxesLabel -> {"x", "y"}, AxesOrigin -> {0, 0} ]
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
Data = [[1, 1.93], [1.1, 1.61], [1.2, 2.27], [1.3, 3.19], [1.4, 3.19], [1.5, 3.71], [1.6, 4.29], [1.7, 4.95], [1.8, 6.07], [1.9, 7.48], [2, 8.72], [2.1, 9.34], [2.2, 11.62]]
def f(x, a, b): return a*x**b
coeff, covariance = curve_fit(f, [point[0] for point in Data],
[point[1] for point in Data])
print("coeff: ",coeff)
x_val = np.arange(1,2.2,0.01)
plt.title('%.5fx**(%.5f)' % tuple(coeff))
plt.plot(x_val, f(x_val, coeff[0], coeff[1]))
plt.scatter([point[0] for point in Data], [point[1] for point in Data], c='k')
plt.xlabel("x"); plt.ylabel("y")
plt.grid(); plt.show()
# R squared
x = np.array([point[0] for point in Data])
y = np.array([point[1] for point in Data])
y_fit = f(x, coeff[0], coeff[1])
ss_res = np.sum((y - y_fit)**2)
ss_tot = np.sum((y - np.mean(y))**2)
r2 = 1 - (ss_res / ss_tot)
print("R Squared: ",r2)
The following link provides the MATLAB codes for implementing the Linearization of nonlinear power model.

Muy buen articulo, fue de mucha ayuda
Thank you